One. Experimental Principle
Real time fluorescence quantitative PCR technology refers to the method of adding fluorescent groups to the PCR reaction system, using fluorescence signal accumulation to monitor the whole PCR process in real time, and finally, the background expression of each sample is determined by relative quantitative or absolute quantitative methods.
Related Application
1. Clinical disease diagnosis
Diagnosis and efficacy evaluation of infectious diseases such as hepatitis, AIDS, avian influenza, tuberculosis and sexually transmitted diseases; Detection of eugenics for the dieases such as thalassemia, hemophilia, abnormal gender development, mental retardation syndrome and fetal malformation; Diagnosis of tumors by detecting the tumor markers and tumor genes; Diagnosis of genetic diseases by detecting genetic genes.
2. Animal disease detection
Detection of avian influenza, Newcastle disease, foot-and-mouth disease, classical swine fever, Salmonella, Escherichia coli, Actinobacillus pleuropneumoniae, parasitic diseases, Bacillus anthracis.
3. Food safety
Detection of food borne microorganisms, food allergens, genetically modified organisms and Enterobacter sakazakii in dairy enterprises.
4. Scientific research
Research on medicine, agriculture and animal husbandry, and quantitative research on biological related molecular biology.
5. Application industry
Medical institutions at all levels, universities and research institutes, CDC, inspection and quarantine bureaus, veterinary stations, food enterprises and dairy factories, etc.
Because qPCR is a real-time quantitative detection of pathogenic pathogen gene nucleic acid, it has more unique advantages than immunological methods such as chemiluminescence, time-resolved and protein chip.
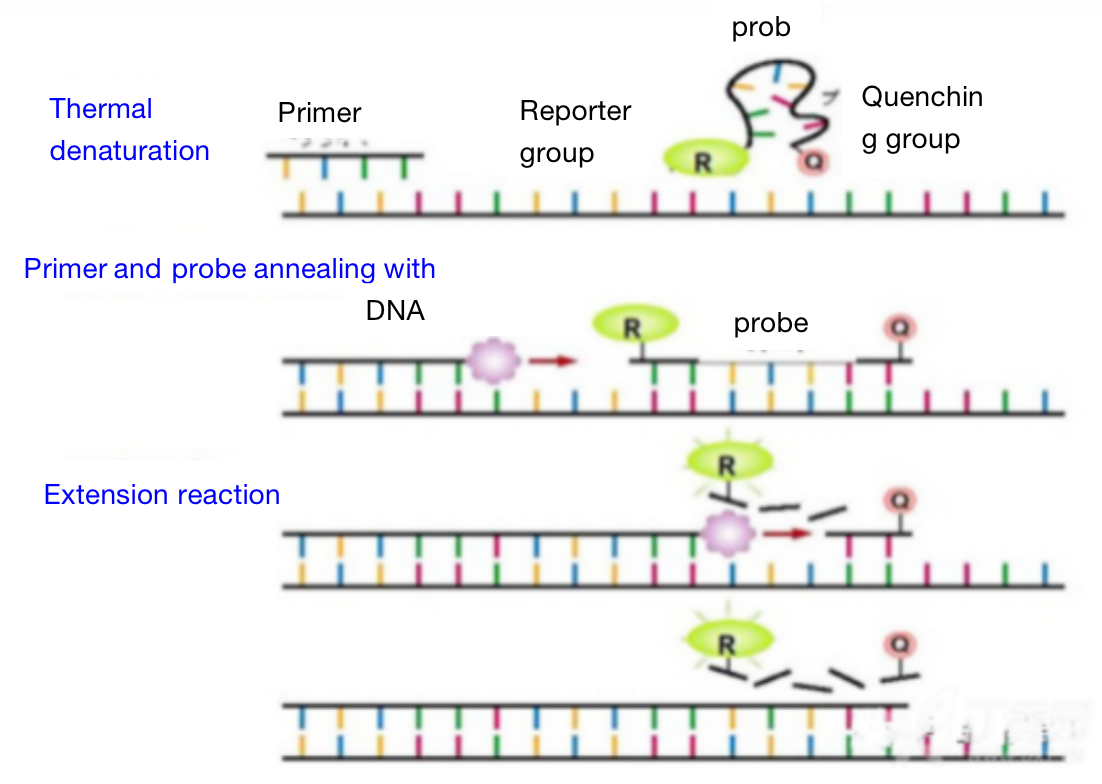
Three. Experimental Method

Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
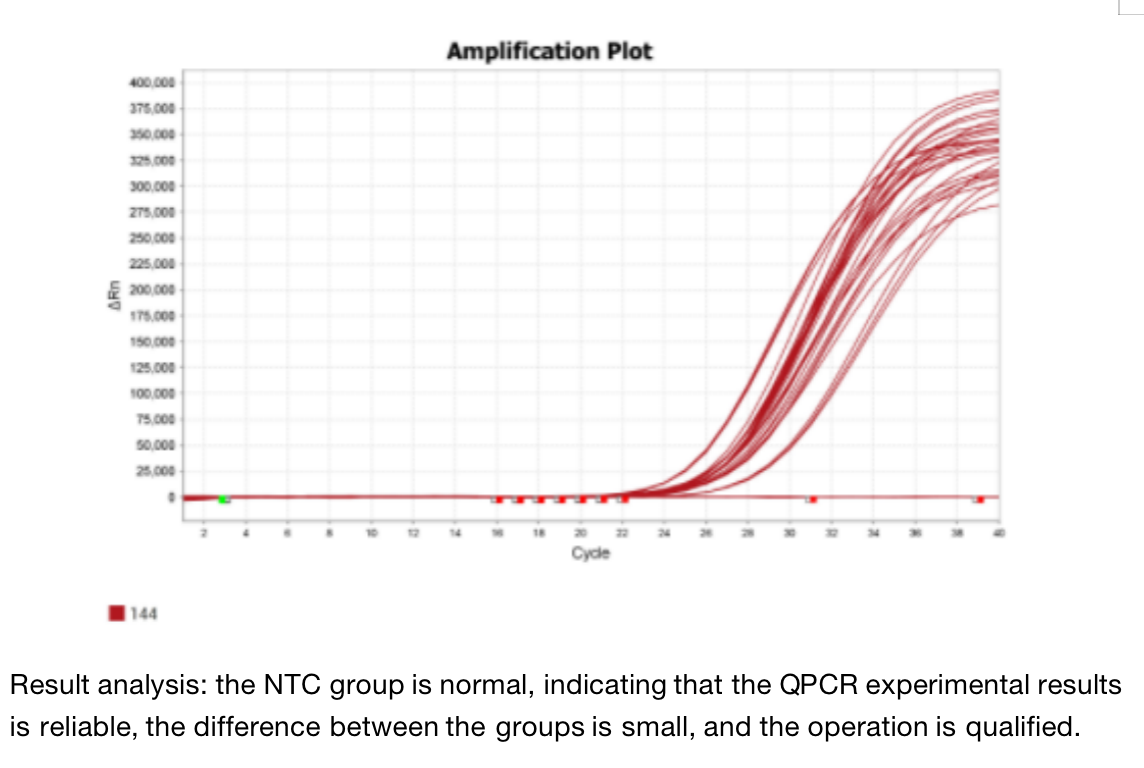
Five. Case Display
Six. Common Problems
1) Primer design
1. Sequence selection should be in the conserved region of the gene;
2. Taqman probe technology requires that the fragment length is 50bp-150bp;
3. Avoid the formation of circular hairpin structure by the primer itself. Avoid forming 4 or more continuous pairings between primers themselves or with primers;
4. Typical primers are 18 to 24 nucleosides long. Primers need to be long enough to ensure sequence uniqueness and reduce the possibility that the sequence exists in non target sequence sites. However, primers longer than 24 nucleosides do not represent higher specificity. Longer sequences may hybridize with wrong paired sequences, which reduces the specificity. And the hybridization of longer sequences is slower than that of short sequence, reducing the yield. TM value is 55-65 ℃, GC content is 40% - 60%;
5. The difference of TM value between primers should not exceed 2 ° C;
6. The 3 'end of the primer should avoid 3 or more consecutive identical bases;
7. In order to avoid genome amplification, primer design should better span two exons;
8. The probe position should be as close to the upstream primer as possible;
9. Base G should be avoided at the 5 'end of the probe and base A should be avoided at the 3' end of the primer. The probe length is usually 25 ~ 35bp, the TM value is 65 ~ 70 ° C, which is usually 5 ~ 10 ° C higher than the primer TM, and the GC content is 40% ~ 70%;
10. In the whole probe, the content of base C was significantly higher than that of base G;
11. In order to ensure the specificity of the primer probe, it is best to verify the designed sequence in blast. If non-specific complementary regions are found, it is recommended to redesign the primer probe.
2) Hot start
Hot start PCR is one of the most important methods to improve the specificity of PCR in addition to good primer design.
3) Magnesium ion concentration
Magnesium ion affects many aspects of PCR, such as affecting the activity of DNA polymerase and further impact the yield; affecting on primer annealing and then further affect specificity. If dNTP and template are combined with magnesium ions, the amount of free ions required for enzyme activity is reduced.
4) Template quality
The quality of the template will affect the output. There may be a variety of contaminants in DNA samples that inhibit PCR.
5) Avoid residual contamination
1. PCR is susceptible to contamination because it is a sensitive amplification technique. A small amount of exogenous DNA contamination can be amplified together with the target template. When the former amplification products are used for new amplification reactions, common source pollution will occur. This is called residual pollution. DNA purified from other samples or cloned DNA can also be a source of contamination (non residual contamination).
2. Good experimental steps can be used in the PCR process to reduce residual contamination. Set up isolated areas for PCR sample preparation and post amplification analysis, and change gloves before carrying out new reactions. Always use negative controls that do not contain templates to detect contamination. Use premixed reaction components instead of adding each reagent of each reaction separately.
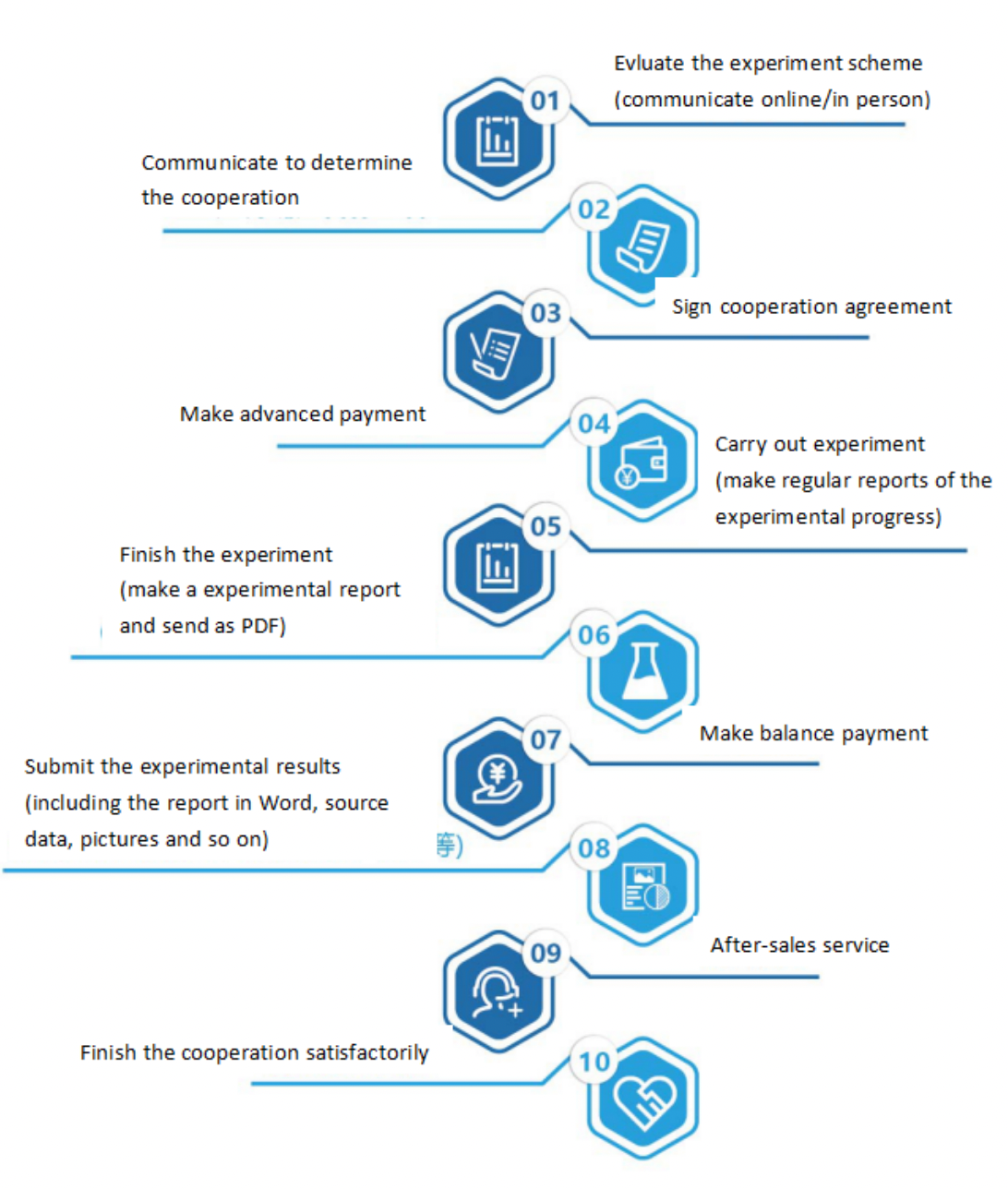
Seven. Service Process