One. Experimental Principle
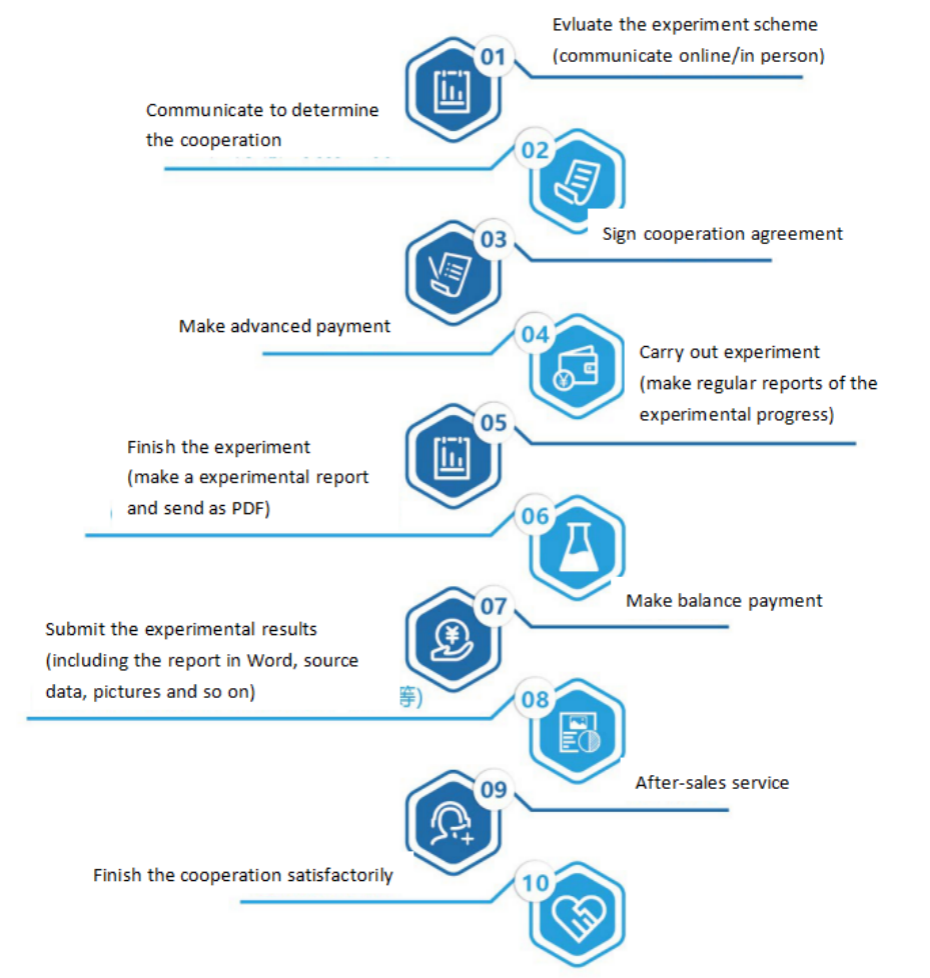
Gel mobility shift assay or electrophoretic mobility shift assay (EMSA) is a technique for studying the interaction between DNA binding proteins and their associated DNA binding sequences, which can be used for qualitative and quantitative analysis. Originally used to study DNA binding proteins, this technique has been used to study the interaction between RNA binding proteins and specific RNA sequences.
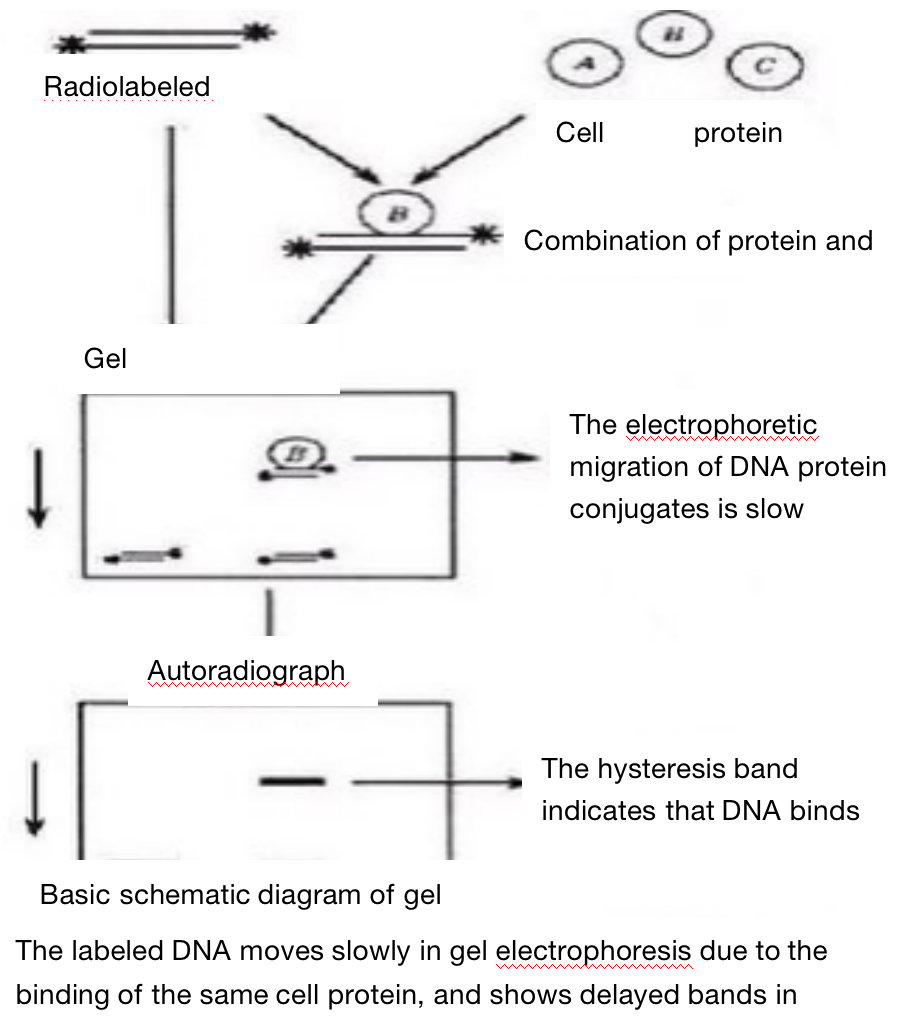
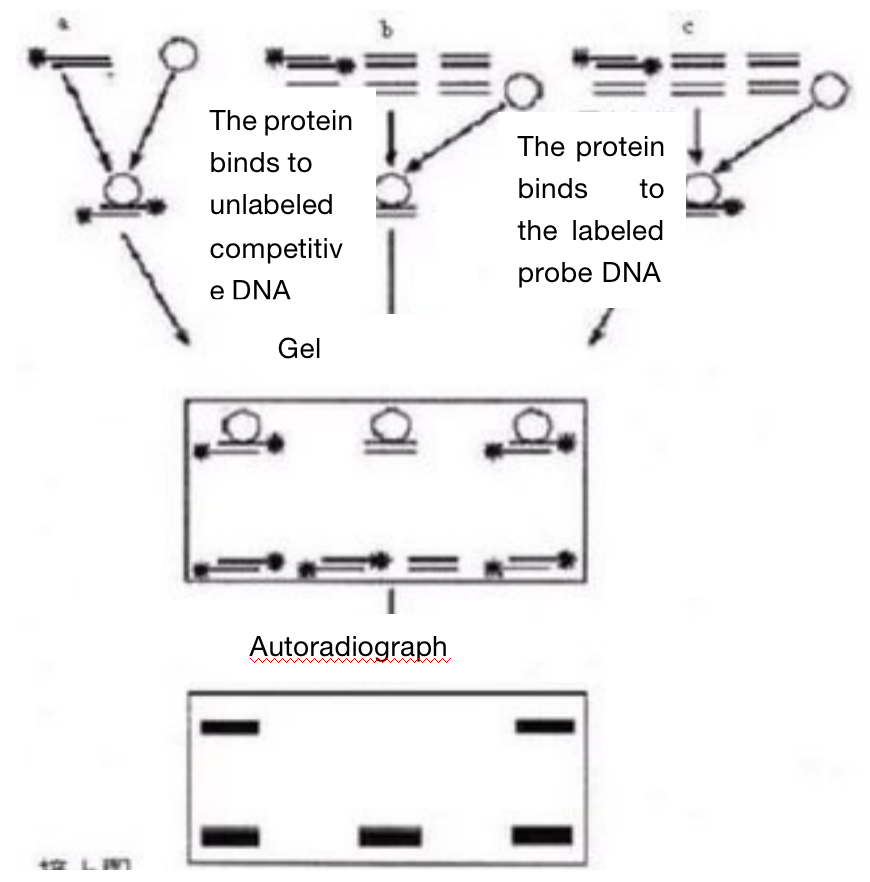
Usually, the purified protein and cell crude extract are kept warm together with 32P isotope labeled DNA or RNA probes, and the complexes and non binding probes are separated on non denaturing polypropylene gel electrophoresis. DNA complexes or RNA complexes move more slowly than unbound probes. Isotope labeled probes can be double stranded or single stranded depending on the binding protein studied. When detecting DNA binding proteins such as transcription regulators, purified proteins, partially purified proteins or nuclear cell extracts can be used. When detecting RNA binding protein, purified or partially purified protein or nuclear or cytoplasmic cell extract can be used according to the location of target RNA binding protein. In the competition experiment, DNA or RNA fragments and oligonucleotide fragments containing protein binding sequences (specific) and other unrelated fragments (nonspecific) are used to determine the specificity of DNA or RNA binding proteins. In the presence of competition between specific and non-specific fragments, the specific binding is determined according to the characteristics and strength of the complex.
Two. Application Introduction
1. Study the interaction between DNA binding protein and its related DNA binding sequence;
2. It can be used for qualitative and quantitative analysis of DNA;
3. Study the interaction between RNA binding proteins and specific RNA sequences.
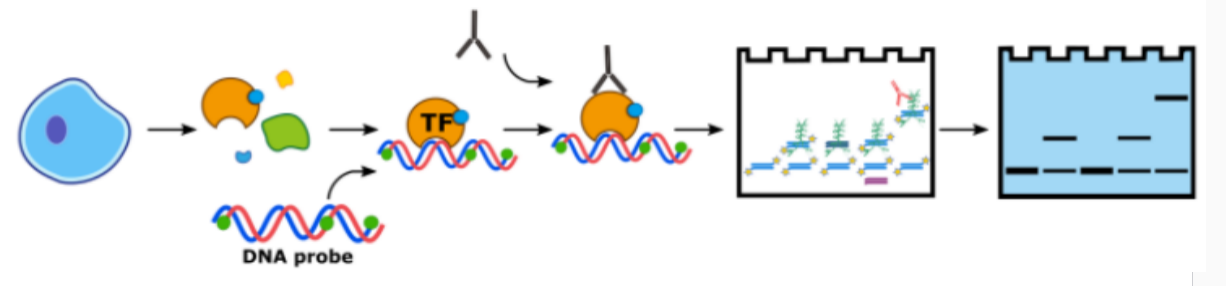
Three. Experimental Method
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin- embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
Five. Case Display
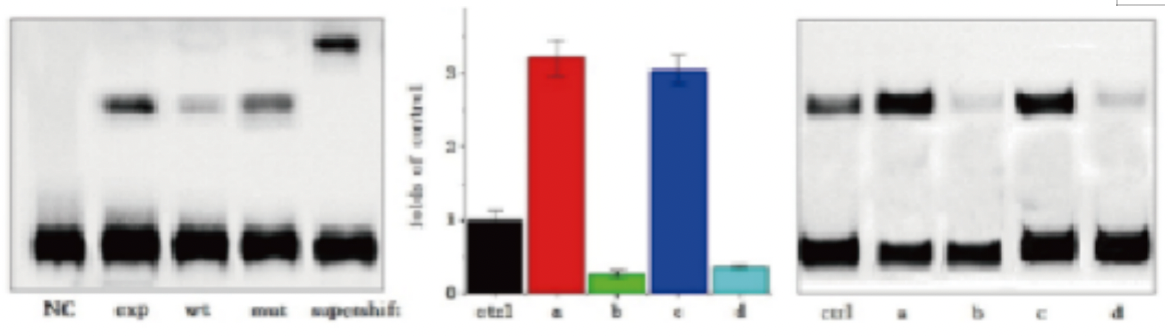
Six. Common Problems
2. The gel mobility experiment is simple and fast in theory, but in order to successfully carry out the gel mobility experiment, some parameters need to be optimized, which is mainly affected by the source of binding proteins and the characteristics of probe binding sites.
3. An important factor to be considered in the selection of DNA probe is that the length of target DNA should be less than 300bp, which is conducive to the electrophoretic separation of non binding probe and protein DNA complex. Double stranded synthetic oligonucleotides and restriction fragments can be used as probes in gel mobility experiments.
3. What complexes can be formed with the following transcription regulators and HeLa nuclear extract: AP1, ap2, CREB, NFkB, Oct1, SP1, TFIID, TFIIB.
4. When HeLa nuclear extract is used as the source of binding protein, each transcription regulator binds with its related DNA homologous sequence to form a characteristic binding form. The following text describes each individual transcription regulator, including the recognized homologous sequence, the size of the factor, specific binding conditions, and the number of complexes that can be formed with HeLa nuclear extract.
Seven. Service Process