One. Experiment Principle
Chromosome immunocoprecipitation is a method based on in vivo analysis, also known as binding site analysis. In the past decade, it has become the main method of epigenetic information research. This technique helps researchers determine what histone modifications occur at a specific location in the genome in the nucleus. CHIP can not only detect the dynamic interaction between trans factors and DNA in vivo, but can also be used to study the relationship between various covalent modifications of histones and gene expression.
The basic principle is to fix the protein-DNA complex in the state of living cells and cut it randomly into small chromatin fragments within a certain length range, then precipitate the complex by immunological methods, specifically enrich the DNA fragments bound to the target protein, and obtain the information of the interaction between protein and DNA through the purification and detection of the target fragment. CHIP can not only detect the dynamic interaction between trans factors and DNA in vivo, but also study the relationship between various covalent modifications of histones and gene expression.
Two. Application Introduction
Nucleic acid and protein are the main biological macromolecules that make up life. Their interaction is the basis of life activities such as growth, reproduction, movement, heredity and metabolism. Studying the interaction between protein and nucleic acid is the key to explore the mystery of life phenomenon. CHIP detection experiment is an important reference for the interaction between protein and nucleic acid.
Chromatin immunocoprecipitation can be applied to: (1) antibodies to histone modifying enzymes as "biomarkers"; (2) transcriptional regulation analysis; (3) drug development and research; (4) DNA loss and apoptosis analysis.
Three. Experimental Methods
Treat the cells with formaldehyde - collect cells, fragment cells with ultrasonic - add antibodies to the target protein, bind to the target protein-DNA complex - add protein A, bind to the antibody-target protein-DNA complex, and precipitate - clean the precipitated complex, remove some nonspecific binding - elute to obtain enriched target protein-DNA complex - de-crosslink, purificate the enriched DNA fragments - PCR analysis.
Four. Sample Delivery Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
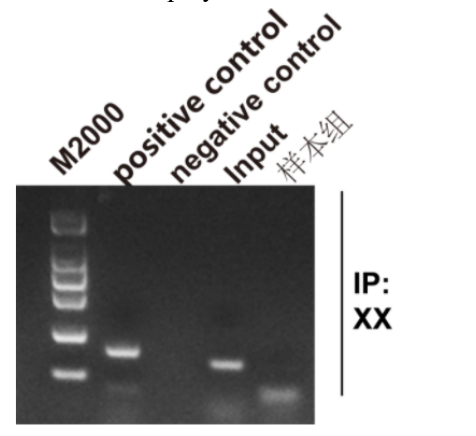
Five. Case Display
Six. Common Problems
1. The concentration of digested chromatin is too low.
2. Chromatin digestion was insufficient and the fragment was too large (greater than 900 bp).
3. There were no products or few products in the PCR reaction of the sample input control group.
4. There was no product in the PCR reaction of positive control histone H3-IP RPL30.
5. The number of products in the negative control Rabbit IgG-IP and positive control histone H3-IP PCR reaction is equal.
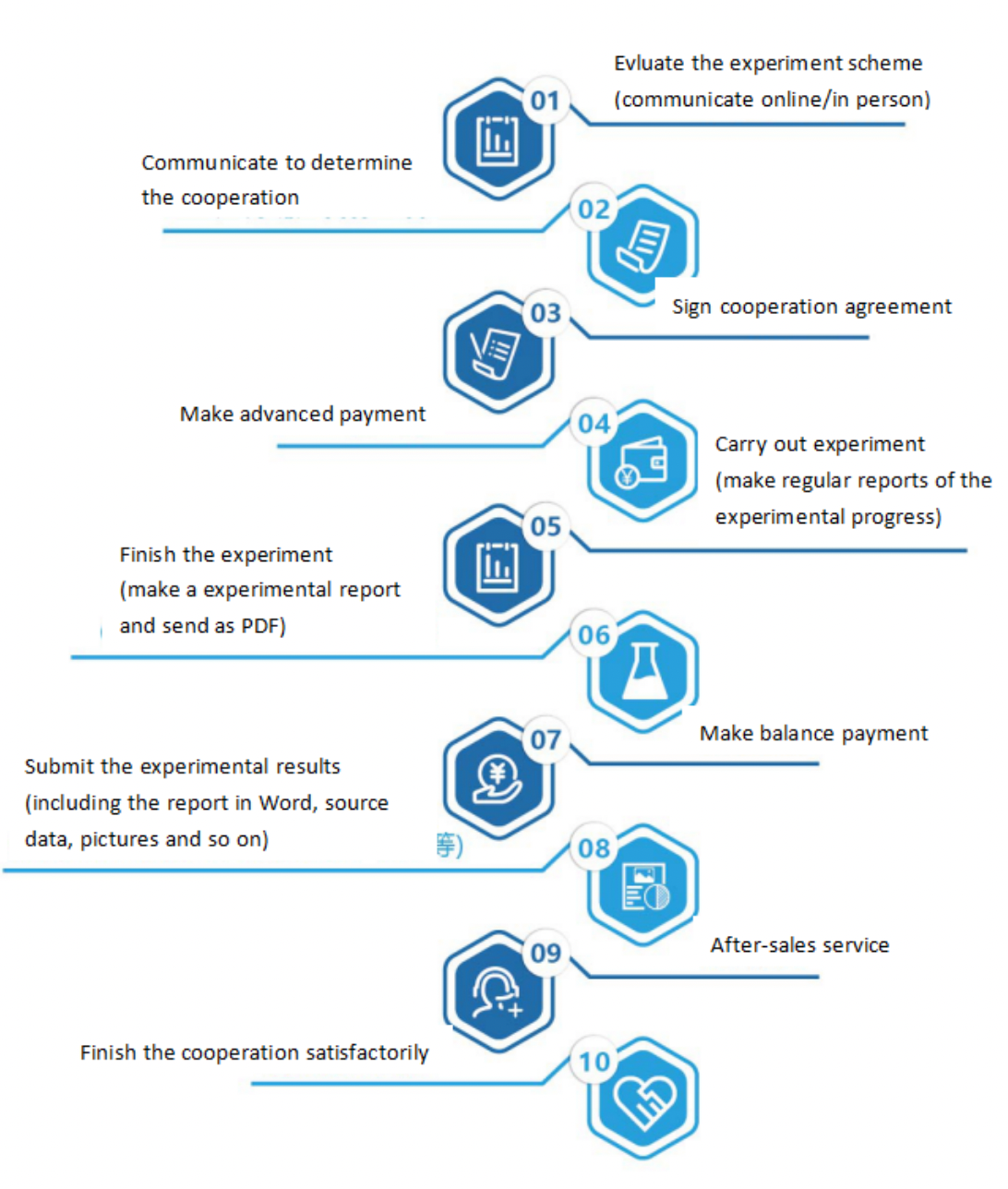
Seven. Service Process