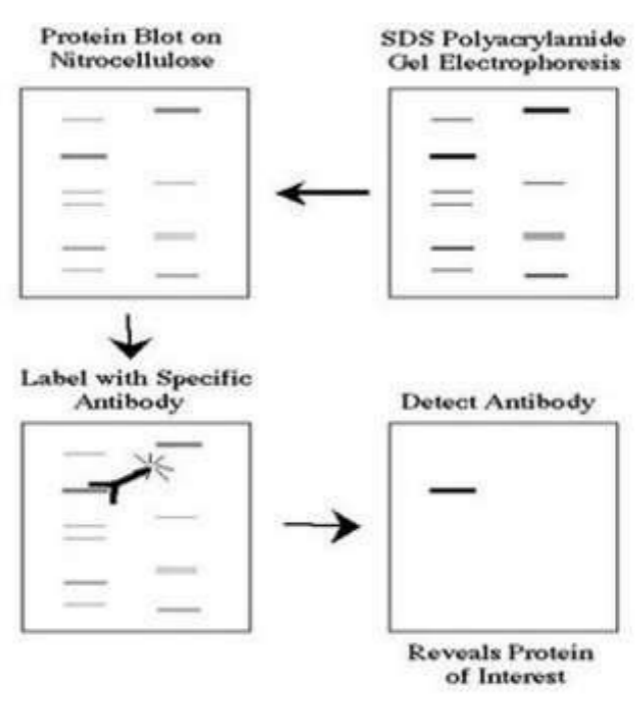
One. Experimental Principle
Western blot adopts polyacrylamide gel electrophoresis, the detected object is protein, "probe" is antibody, and "color development" is marked with second antibody. The protein sample separated by PAGE is transferred to the solid support (such as nitrocellulose membrane NC membrane). The solid support adsorbs the protein in the form of non covalent bond, and can keep the type of polypeptide separated by electrophoresis and its biological activity unchanged. Take the protein or polypeptide on the solid support as the antigen, react with the corresponding antibody, then react with the enzyme or isotope labeled second antibody, and detect the protein components expressed by the specific target gene separated by electrophoresis through substrate color development or autoradiography. This technique is also widely used to detect the expression of protein level. The diagram is as foolow:
Western blot color development methods mainly include the following:
1. autoradiography
2. Substrate chemiluminescence ECL
3. substrate fluorescence ECF
4. substrate DAB color development.
At present, substrate chemiluminescence ECL and substrate DAB color development are commonly used. The first method can demonstrate the level and experimental conditions. At present, substrate chemiluminescence ECL is usually used in published articles. It can be done as long as you buy a ready-made kit, the operation is also relatively simple. The principle is as follows (the second antibody is marked with HRP): the reaction substrate is peroxide + luminol. In case of HRP, it emits light, which can expose the film and wash out the strip.
Two. Application Introduction
Western blot uses an experimental method which is often used in molecular biology, biochemistry and immunogenetics, and it is a qualitative and semi quantitative analysis method of protein. It is to stain the cells or biological tissue samples treated by gel electrophoresis with specific antibodies, and obtain the information about the expression of specific proteins in the analyzed cells or tissues by analyzing the location and depth of staining.
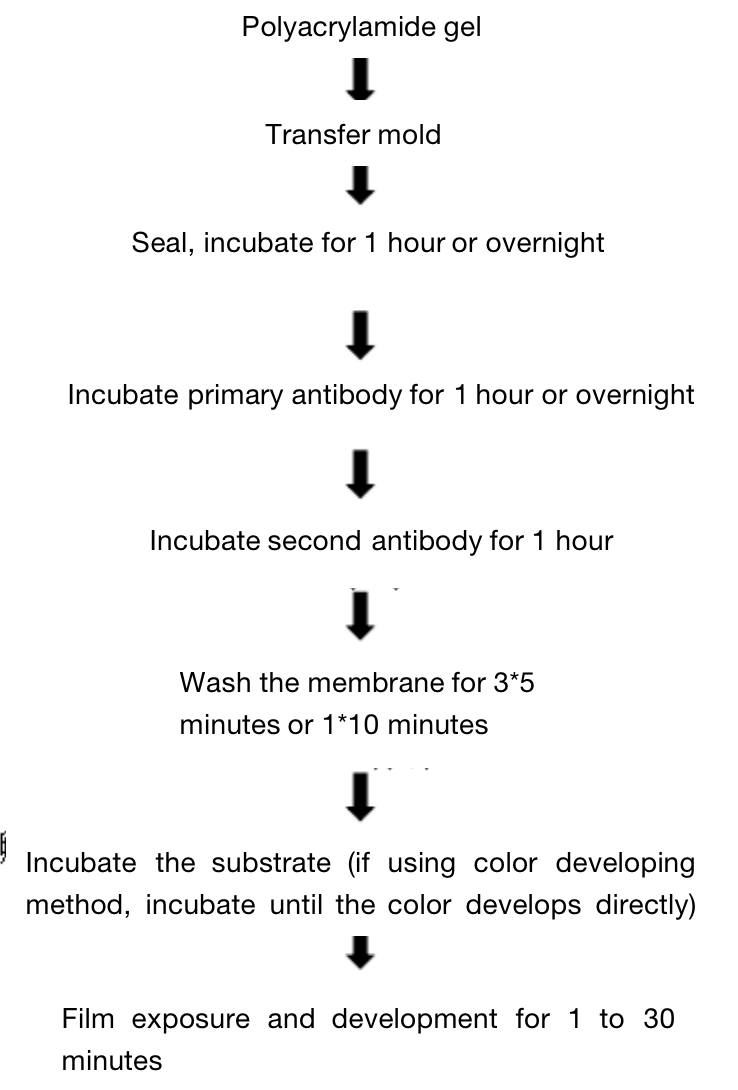
Three. Experimental Method
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
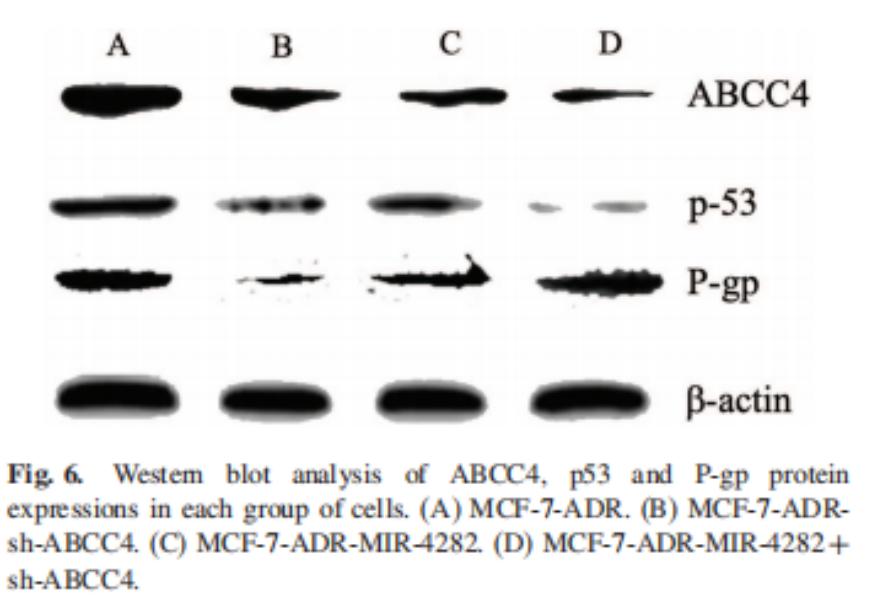
Five. Case Display
Six.Common Problems
Problems | Possible Cause | Verification or Solution |
High background | Incomplete sealing | Proplong the sealing time, replace with an appropriate sealant (skimmed milk powder, BSA, serum, etc.) |
The concentration of the primary antibody is too high. | Increase the dilution multiple of primary antibody | |
The temperature for antibody incubation is too high. | Incubate at 4°C | |
The second antibody is nonspecific binding or cross react with blocker | Set up second antibody control (without primary antibody),reduce the concentration of secondary antibody | |
Incomplete washing of membrane | Increase washing times | |
The membrane is inappropriate | The background of NC membrane is lower than that of PVDF membrane | |
The membrane is dried | Ensure the reaction liquid is sufficient to avoid dry film | |
No positive bands or they are very weak | The primary antibody and second antibody do not match | When ordering reagents, carefully select the antibodies, and substrates matching between primary antibody and tissue species, primary antibody and secondary antibody or / and substrate and enzyme system. The effectiveness of the secondary detection system can be verified by setting internal reference. |
The concentration of the primary antibody or second antibody is low. | Increase the concentration of the antibody, prolong the incubation time | |
There is cross reaction between sealant and primary antibody or second antibody. | When sealing, use mild detergent or replace the sealing agent (commonly used skimmed milk, BSA, serum or gelatin) | |
The primary antibody does not recognize the target protein of the target species. | Check the instructions, or make ClustalW comparison, and set a positive control | |
There is no target protein in the sample or the content of target protein is too low (antigen is invalid) | Set a positive control. If the positive control has results but the sample does not, it may be that the sample does not contain target protein or the content of target protein is too low. The latter can increase the sample loading amount on the sample by at least 20-30 micrograms of protein per well, use protease inhibitor during sample preparation, or extract the target protein by grading. | |
The transfer of membrane is incomplete or the washing of membrane is excessive. | Use Ponceau S to detect the membrane transfer effect. The PVDF film needs to be soaked, and the film transfer operation needs to be correct. Do not wash the membrane excessively. | |
Over-sealed | Use the antibody diluent containing 0.5% skimmed milk or milk without lotus root juice, or change the sealing agent to reduce the sealing time | |
The primary antibody is invalid. | Use the antibody within the validity period, store separately, avoid repeated freezing and thawing. The working solution should be prepared when using. | |
The second antibody is inhibited by sodium azide. | Avoid sodium azide (an inhibitor of HRP) in all solutions and containers | |
The enzyme and substrate are invalid. | Directly mix the enzyme with the substrate. If it does not develop color, it indicates that the enzyme is inactivated. Select the active enzyme linked substances within the validity period and use fresh substrates | |
The exposure time is too short. | Prolong the exposure time | |
Multiple nonspecific bands or bands in wrong positions | The excessive number of cell passages leads to the differentiation of protein expression pattern | Use original or less passage cell lines, or parallel experiments |
The protein samples expressed in vivo have a variety of modification forms: acetylation, phosphorylation, methylation, alkylation, glycosylation and so on | Check the literature and use reagent that can de-modify to restore the protein to its correct size. | |
The protein sample is degraded | Use freshly prepared samples and protease inhibitors. | |
New proteins or homologous proteins share different splicing modes of the same expression | Check other literature reports, or do BLAST search, use cell lines or tissues reported in the instruction manual. | |
The concentration of the primary antibody is too high. | Reducing antibody concentration can reduce nonspecific bands | |
The concentration of the second antibody is too high, lead to nonspecific binding. | Reduce the antibody concentration, increase the second antibody control, select the second antibody with stronger specificity and only target the heavy chain | |
The antibody is not purified. | Use monoclonal or affinity purified antibodies to reduce nonspecific bands | |
There are dimers or multimers in the protein. | Before SDS-PAGE electrophoresis, boil for ten minutes tofacilitate protein depolymerization and denaturation | |
Background has white/black spots | There are bubbles or uneven distribution of antibodies during membrane transfer | Remove bubbles as much as possible and keep shaking during antibody incubation |
Antibodies bind with sealer | Filter sealer. | |
White bands on dark film | Primary or second antibodies are over-added. | Dilute the concentration of the antibodies. |
Targets bands are too low/high | The concentration of SDS-PAGE gel is inappropritate | Adjust the gel concentration. Low concentration gel is used for high molecular weight protein and high concentration gel is used for high molecular weight protein |
“Smile” bands | The transfer is too fast, or the electrophoresis temperature is too high. | Reduce electrophoresis speed, perform electrophoresis at low temperature (cold chamber) |
Incomplete membrane transfer | The membrane is not completely and evenly wet | Use 100% methanol to soak the membrane |
The membrane is not completely wet | Select the membrane with small pore size to shorten the transfer time | |
The isoelectric point of the target protein is equal to or close to the pH value of the transfer buffer. | Try using other buffers such as caps buffer (pH = 10.5) or low pH buffer such as acetic acid buffer | |
The concentration of methanol is too high | Too high methanol concentration will lead to the separation of proteins from SDS, which will precipitate in the gel. At the same time, it will shrink or harden the gel, inhibiting the transfer of high molecular weight proteins. It can be solved by reducing the concentration of methanol or use ethanol or isopropanol instead |
Other problems
(1) The molecular weight of protein is high or low. It may be that the concentration of gel does not correspond to the concentration of target protein.
(2) Protein degradation. After protein degradation, it is likely that the main band will appear at a place lower than the original position, and then some other bands will appear. The main feature is that all bands are lower than normal, and the bands are blurred and unclear.
(3) All the strips are connected into one piece without interval. The most likely reason is excessive sample loading, followed by sample dispersion (for example, Electrophoresis stopped for a long time leads to sample dispersion).
(4) The whole band is in a "︶" shape: the gel is not uniformly cooled, and the electrophoresis tank is aging.
(5) The whole band looks like " ︵": the left and right ends of the gel are not solidify well.
(6) Bromophenol blue tailing: the sample does not dissolve well.
(7) Longitudinal texture: there are insoluble particles exist in the sample.
(8) Bromophenol blue is very thick: the concentration effect of concentrated gel is not good. It may be that the time for concentration is too short or the concentrated gel is mismatched.
(9) Can't move in the separating gel: Tris-HCl pH value is wrong, or the SDS is forgot to add.
Seven. Sevice Process