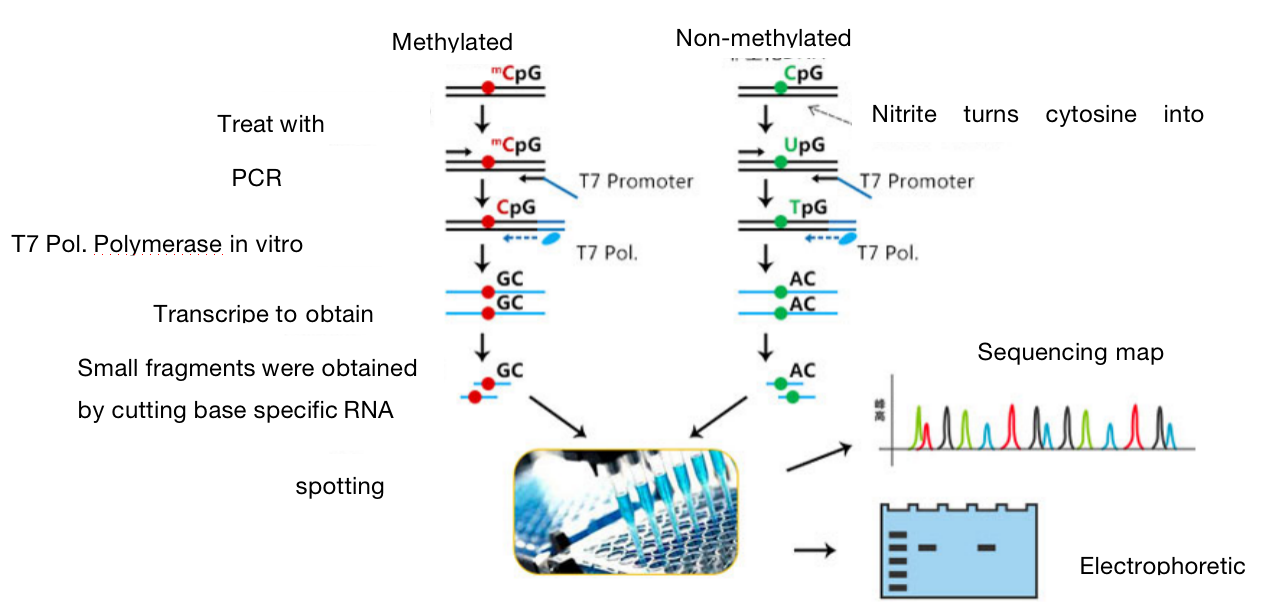
One. Experimental Principle
DNA methylation exists in most eukaryotes. BSP methylation sequences through bisulfite treatment of genomic DNA, unmethylated cytosine is deaminated and transformed into uracil. In the subsequent PCR reaction, uracil is transformed into thymine, while methylated cytosine cannot be deaminated and is retained at the completion of the reaction. We cloned and sequenced the amplified PCR products by TA, the specific situation of methylation can be analyzed. This method has high accuracy and is the gold standard for methylation detection.
Two. Application Introduction
In the development of malignant tumors, the state of methylation is not invariable. The degree of genome-wide hypomethylation in tumor cells is closely related to disease progression, tumor size and malignancy. DNA methylation detection is of great significance to judge the malignancy of tumors.
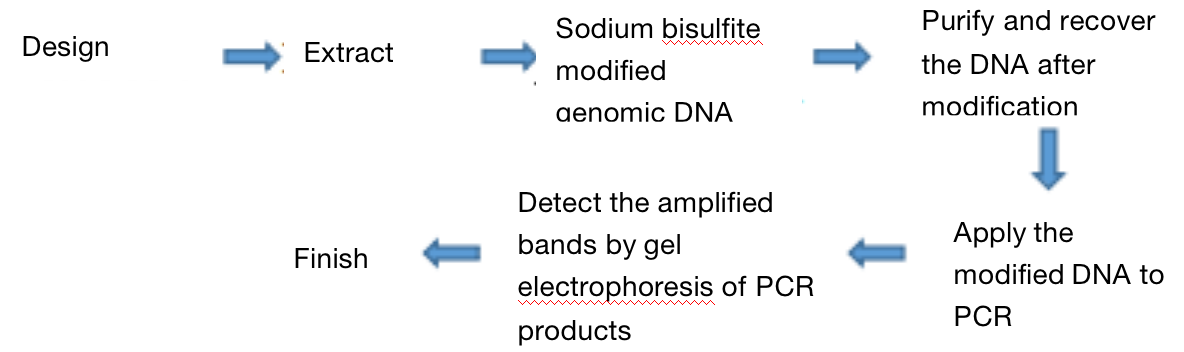
Three. Experimental Methods
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
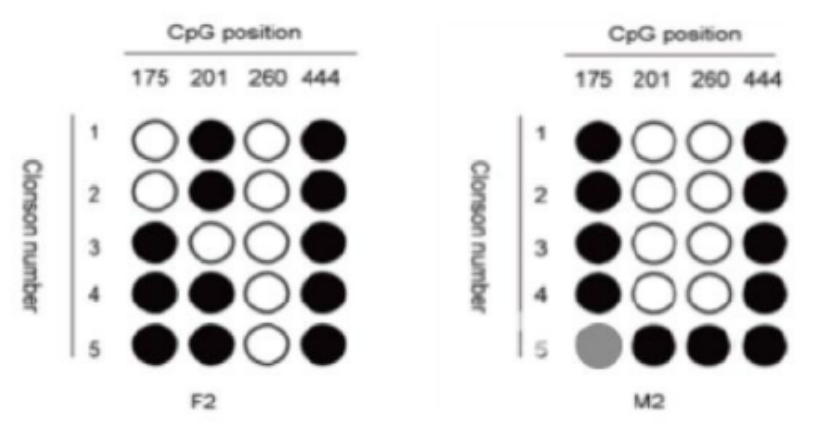
Five. Case Display
Six. Common Problems
1. Protease K can be prepared into 20mg/ml with sterilized double distilled water;
2. RNase must be prepared into RNase without DNA enzyme, that is, it shou;ld be reprocessed after purchasing commercial RNase and prepared into 10mg / ml.
3. There are two methods to verify the purity of extracted DNA: calculating od ratio with ultraviolet spectrophotometer; 2: 1%-1.5% agarose gel electrophoresis. The second method can completely clarify the purity of the genomic DNA and estimate its concentration according to the sample loading amount of marker for further modification.
4. The amount of genomic DNA does not need to be very accurate, rather more than less, because it will be lost in the later purification and recovery steps, and the modification of this method can be up to 4ug.
5. All reagents must be prepared fresh, so the technology of liquid preparation should be qualified, which should be fast and accurate.
6. The sodium bisulfite solution is strongly acidic. The pH must be adjusted to 5.0 with alkali, otherwise the inappropriate pH will affect the subsequent purification and absorption.
7. Water bath is best for 16 hours, although it can be as short as 8 hours, but the latter modification will be incomplete.
8. When using the syringe, be sure to apply force evenly and gently. If violence is used, the film in the small column will be crushed and lose its function.
9. Ammonium acetate and glycogen do not need to be prepared fresh. Glycogen can be stored at - 20 ℃ after preparation and ammonium acetate can be stored at ambient temperature, because such concentration of ammonium acetate is very difficult to dissolve. Once placed at 4 ℃, there will be a lot of solute precipitation when taken out.
10. Isopropanol and 70% ethanol do not need fresh preparation, but if the dosage is large, it is also very convenient to prepare on site. The key to this step is the combination of resin and DNA, which again emphasizes the importance of adjusting sodium sulfite pH in the second part. Because the combination of resin and DNA requires an appropriate pH, if the previous step is not done well, the resin cannot combine well with DNA in this step, which will bring disastrous consequences, that is, DNA is squeezed out with the liquid, and there is actually no DNA when eluting.
11. For agarose gel electrophoresis of PCR products, newly prepared electrophoresis solution should be used. The concentration of gel is 1%-2%.
12. During the recovery of gel DNA, observe the band position under 300nm UV lamp, and cut the gel at the location of the target fragment as small as possible to ensure specificity.
13. Ultraviolet irradiation time should not be too long, otherwise it will damage DNA.
14. Store the the recovered DNAat -20 degrees if don’t use immediately, it is very stable within a few months.
15. The coated plate should be uniform to ensure that Xgar and IPTG are evenly distributed on the plate surface;
16. Do not let the blue and white spots grow too full, otherwise it is easy to select two clones at a time.
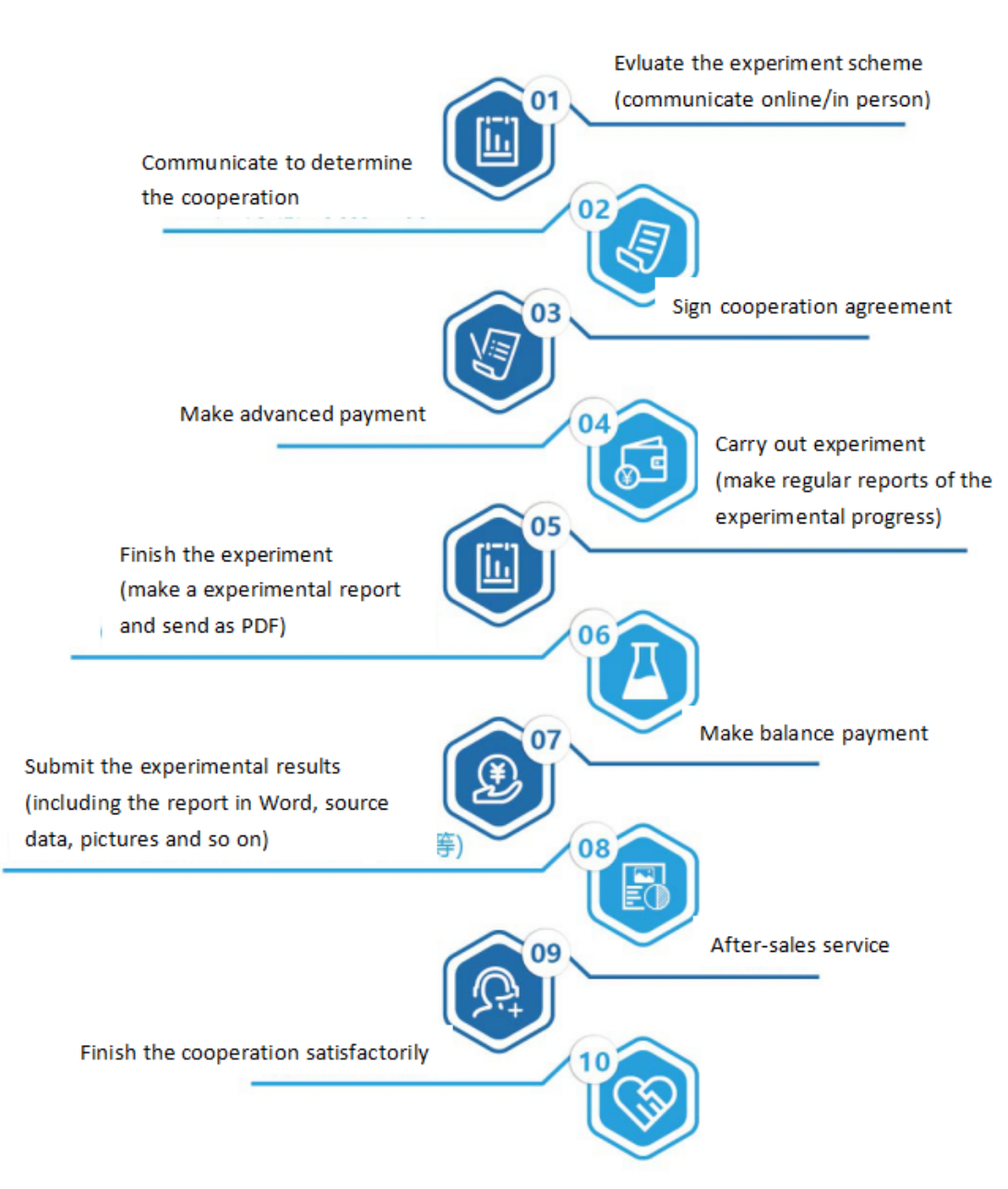
Seven. Service Process