One. Experiment Principle
Short tandem repeats (STR), also known as Microsatellite DNA or simple repeat sequence (SRS or SSR), are nucleotide repeats widely existing in the genomes of prokaryotes and eukaryotes. Replication slippage caused by mismatch between DNA strands during mitosis is considered to be a common cause of STR, and the probability of replication slippage varies according to the size of different replication units and between different species.
STR is formed by connecting core repeat units end to end and repeating for many times. The core sequence structure of each STR is the same, with a length of 2-6bp, but the number of repeat units and the length of repeat regions are different. Therefore, the distribution of STR among different races and populations is different, which constitutes STR genetic polymorphism. Different individuals have different repetition times at a homologous STR locus, so STR locus analysis can also identify individuals, just like fingerprint identification. By identifying the specific sequence repeats of an individual genome at a specific location, its gene file can be created. Based on this, STR analysis has become an important identification and analysis method, which is applied to the fields of forensic medicine, paternity and cell identification.
Two. Application Introduction
Cell lines have been widely used in the study of various diseases because of their unlimited passage and proliferation, in vitro culture and simple culture conditions.
Three. Experiment Method
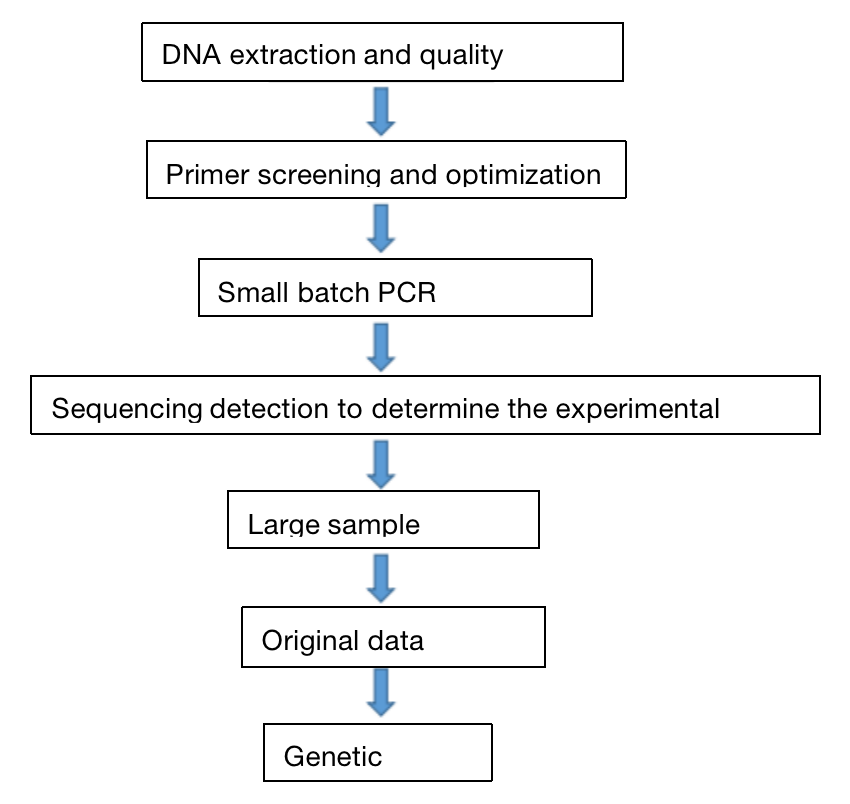
Four. Samples Delivery Requirements
1. Cell suspension (number of cells ≥ 106) - provide cells. After centrifugation with PBS suspension cells, remove the supernatant and transport the cell precipitation with ice bag.
2. Genomic DNA (≥ 20) μl. Concentration ≥ 50ng/μl) - when providing DNA samples, please transport them in ice bags and indicate the DNA content.
3. Only the cell lines with existing STR map data in ATCC and DSMZ databases are identified.
4. STR identified samples are all human derived cells, and 21 gene loci are detected.
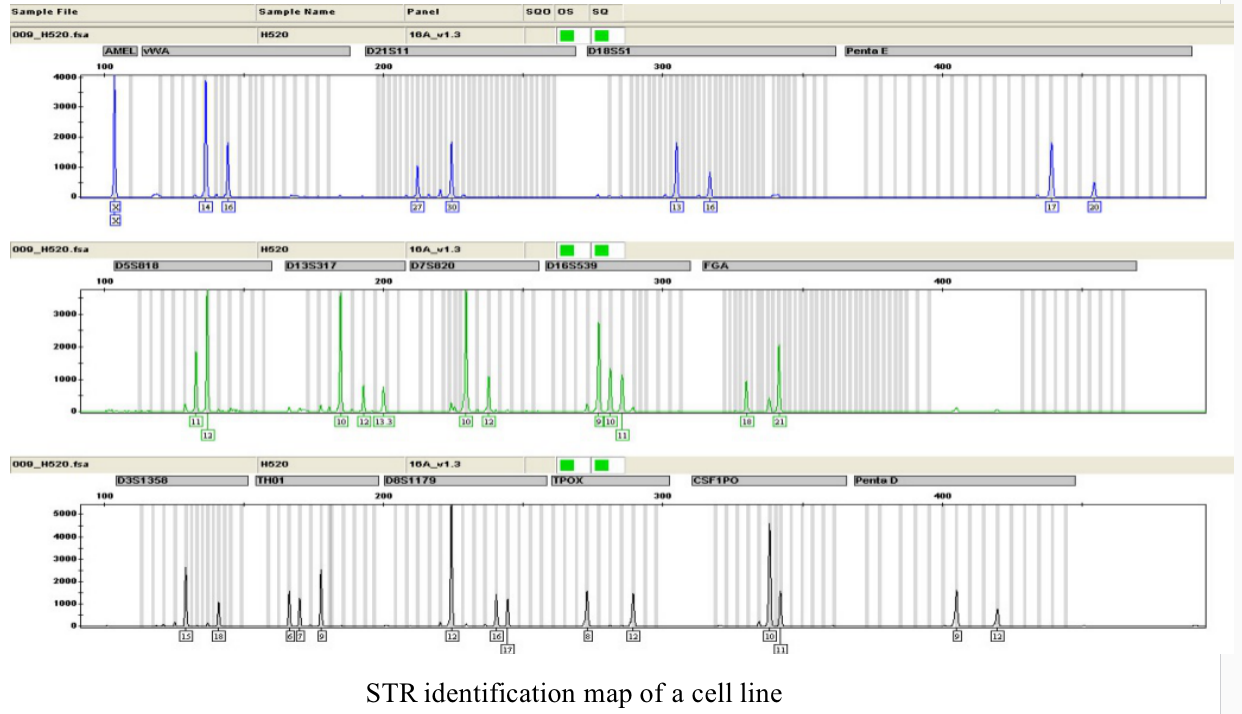
Five. Case Display
Six. Common Problems
1. How to know whether cell lines are cross contaminated
If there is cross contamination between cell lines and there may be more than two cell lines for identification, more than two peaks will appear at multiple sites during STR site detection. The peak height value indicates the signal intensity of the allele. Theoretically, the peak value is directly related to the DNA concentration of the sample. Therefore, in the mixed cell lines with cross contamination, some peaks in one site are significantly higher than other peaks, in which the large peak belongs to the main cell line in the mixed cell line and the small peak belongs to the secondary cell line.
It is worth noting that when two or more cells are mixed, the test results look very much like the "genetic instability" of the cell line -- when there are subpopulations in a cell line, especially some cancer cell lines, the STR characteristics at some loci will be different due to the existence of genetic instability. Therefore, it is very important for experienced molecular experts to help analyze complex electrophoretic maps (STR peaks) to judge whether multiple alleles are caused by cross contamination of cell lines.
2. How to judge the identity of cell lines based on STR data
After receiving the cell line identification results and STR information, it can be compared with the reference database. If each STR locus of the cell sample can coincide with the STR locus of the reference cell, it means that the cell line is correct; otherwise, it means that your cell line may be mislabeled, cross contaminated or genetically unstable. Generally, the cell line is considered correct if the matching degree is ≥ 80%, and the source of the cell line needs to be suspected if the matching degree is < 80%.
If the cell line is identified to have cross contamination or wrong labeling, it is necessary to identify the cells of earlier generations to find the origin of the problem or purchase a new cell line directly from a reliable institution.
3. What if the cell lines don't match the results in the database?
If your cell information is not found in the known database, you can use your own cell genetic information to establish your own genetic characteristics, so that you can compare your own cell bank in the future.
4. Why does the final conclusion in the report fail to match any known data?
The biggest possibility is that the standard sequence of this cell is not included in the international cell bank, resulting in the incompatibility between the submitted sample and any sequence. Most of this is because of the cell line, which may be independently established by the domestic research group.
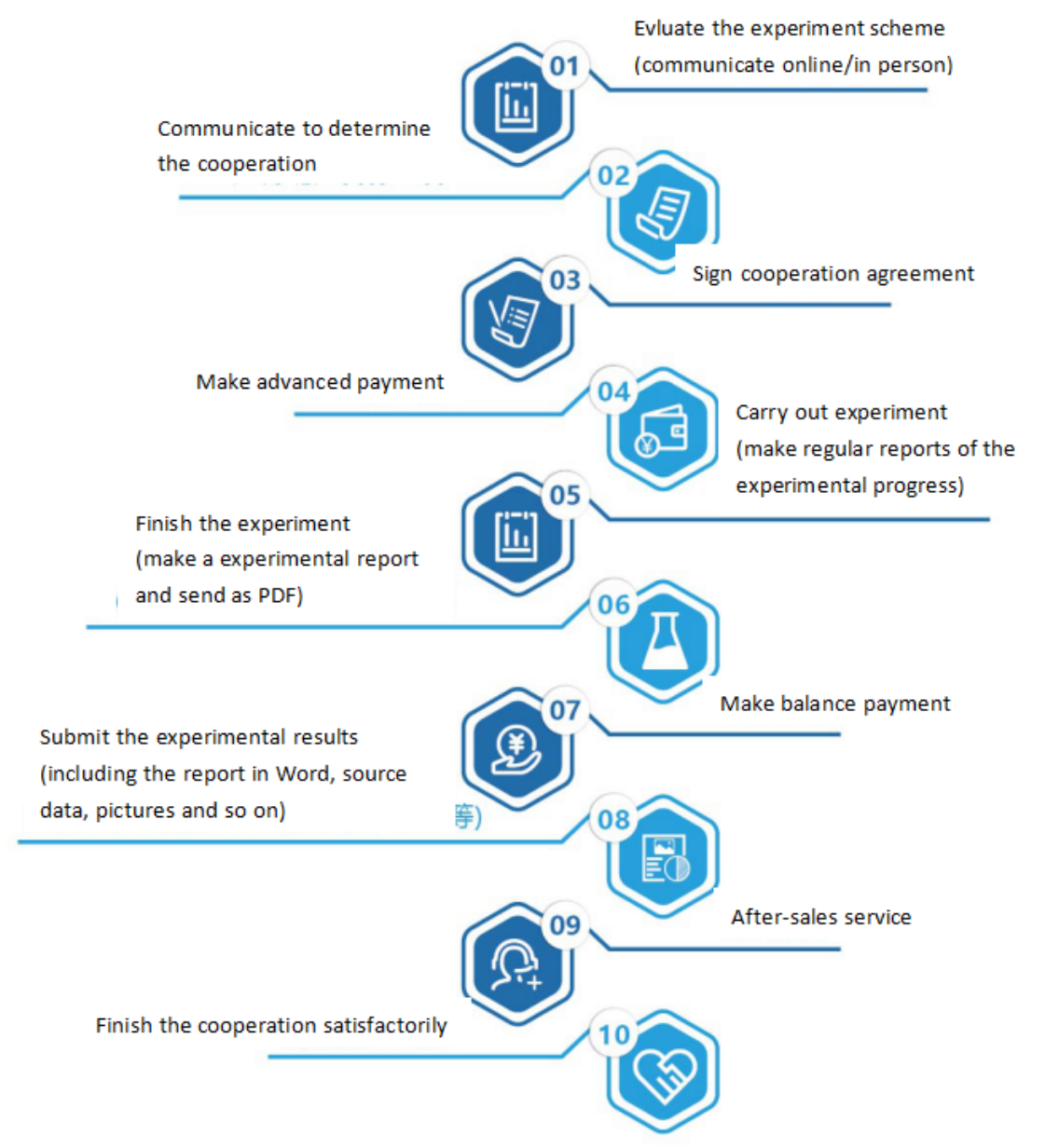
Seven. Service Process