One. Experimental Principle
Southern blot, also known as gel electrophoresis imprint hybridization technology, uses nitric acid fiber membrane, filter paper or nylon membrane with the function of adsorbing DNA. First, DNA fragments are subjected to gel electrophoresis, and the electrophoretic DNA bands are adsorbed on the membrane. Then, hybridization between isotope labeled probe and tested DNA is directly carried out on the membrane. Finally, the hybridization results are detected by autoradiography, so as to detect the specific DNA sequence contained in a DNA sample.
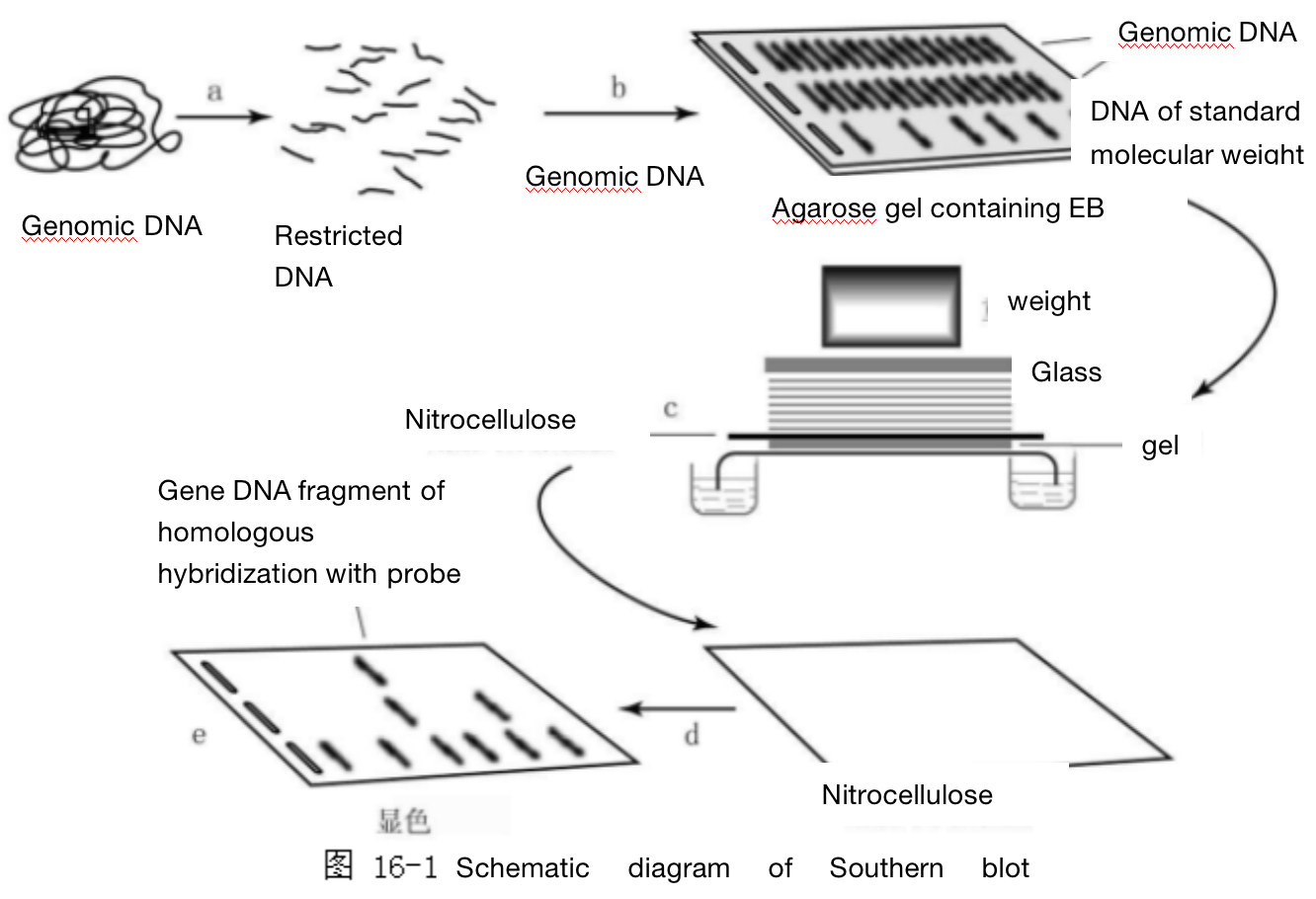
(a) Genomic DNA digested by endonuclease
(b) Separation of DNA fragments by agarose gel electrophoresis
(c) DNA bands in gel is transferred to nitrocellulose membrane
(d) Hybridization between membrane and labeled DNA molecular probe
(e) Chromogenic display of hybridization DNA bands
Two. Application Introduction
Technology application
1. Qualitative and quantitative analysis of specific genes in the genome;
2. Gene mutation analysis and restriction fragment length polymorphism analysis.
Scope of application
1. Genetic disease diagnosis: the traditional gene diagnosis technology is mainly based on gene probe technology to create some detection methods - Southern blot.
2. DNA map analysis: a. high specificity; b. Stable heredity; c. Stable somatic cell
3. Detect DNA and its content in the sample
4. PCR product analysis
Three. Experimental Method
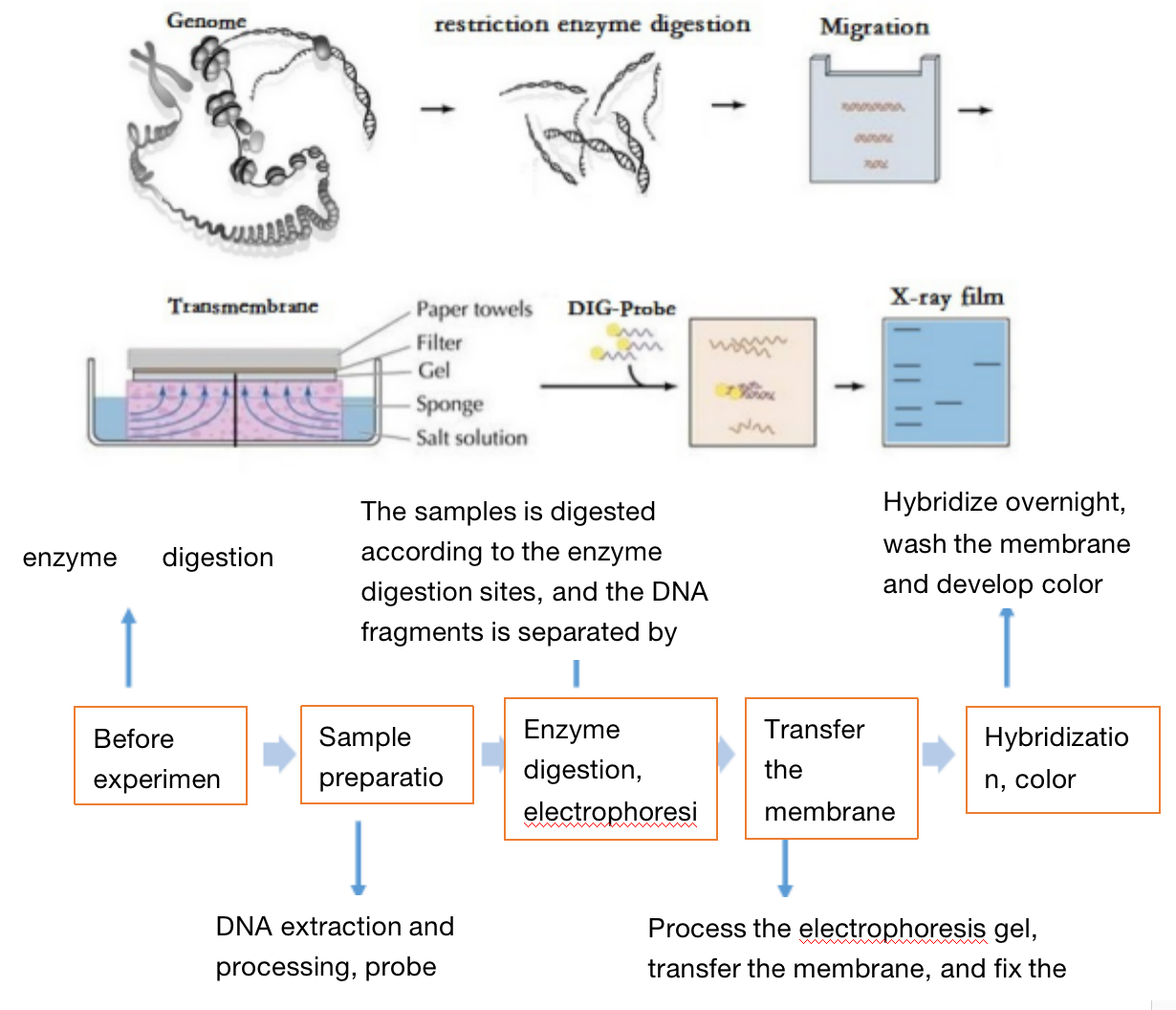
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
Five. Case Display
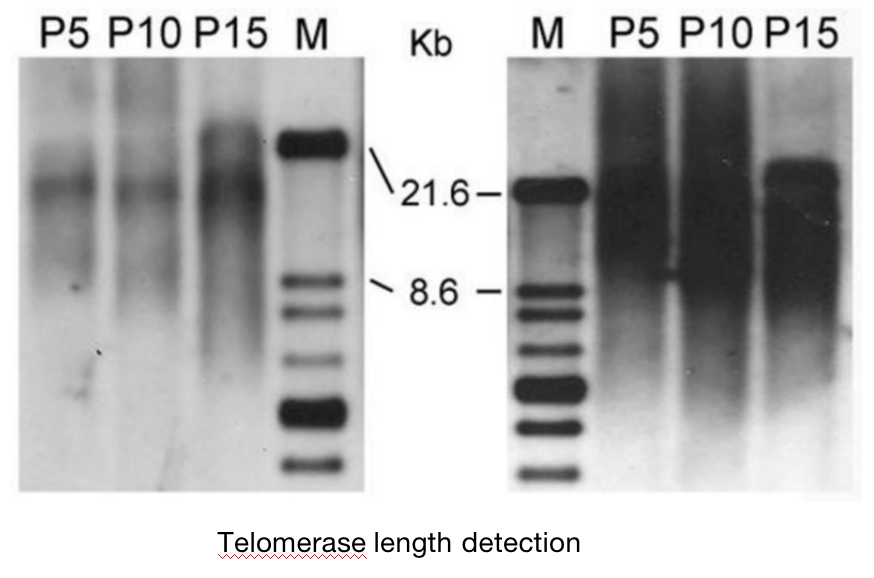
Southern blot has been widely used to analyze DNA and telomere structure. DNA is digested with restriction endonuclease Hinf I or Rsa I, and then fragments of different sizes are separated by agarose electrophoresis and transferred to nitrate fiber or nylon membrane. Telomere specific probes labeled with 32P isotope or biotin and alkaline phosphatase are hybridized with them (CCCATT) n. The terminal restriction fragment (TRF) is quantitatively measured by optical densitometer.
The data obtained by TRF method not only represent the telomere length of all chromosomes, but also include the length of some sub telomere regions. Therefore, TRF method can not provide the actual length of telomeres
Six. Common Problems
1. What’s the effect of nucleic acid quality on Southern hybridization experiment
A: the quality of nucleic acid has a direct impact on the experiment. It requires that the total amount provided by the customer is sufficient -- experimental consumption (loading amount), and it also requires to ensure the quality -- prepare materials as required.
The OD and concentration are generally required by DNA sample as follows:
OD260/280=1.8~2.0,OD260/230>2.0, concentration≥100ng/ul. At the same time, the DNA sample needs to be subjected to agarose gel electrophoresis. The electrophoresis results show that the DNA sample is not polluted by protein, RNA and other substances, there is no degradation dispersion, and the bands are clear.
2. What's probe design and what are the principles?
A: the probe is a digoxigenin labeled DNA sequence homologous to the target gene sequence to be tested, which can be specifically combined with the digested genome fragment. The probe design has the following principles:
(1) Moderate length. The fragment length of the probe is generally controlled at 300-1000 BP. If the probe is too long, the probability of nonspecific binding will increase. If the probe is too short, the probe will bind too few digoxin markers and the luminescence signal will be weakened.
(2) Strong specificity. The best probe sequence is only homologous with the target gene, and the fragment that homologous with the whole genome sequence is less than 30%.
(3) The content of ATGC is uniform, there is no hairpin structure and highly repetitive sequence.
3. How to select restriction enzymes for genome fragmentation?
A: (1) The target gene sequence cannot contain this spot.
(2) The commonly used endonuclease has favorable price and high efficiency
(3) The pre experiment can fragment the genome, and there is no obvious main band in the dispersion state of electrophoretic detection.
(4) Select according to the enzyme digestion sites on the transformation vector
4. How to reduce non-specific exposure bands?
We synthesized a double stranded digoxin labeled probe by PCR and detected whether the probe was labeled successfully by electrophoresis. If the band is single and slightly larger than the control group, it is considered that the probe is synthesized successfully (as shown in Figure 3).
In addition, before synthesizing the probe, the probe sequence needs to be compared on NCBI. If it fail to blast the highly homologous sequences in the species to be tested, the probe is considered to be specific.

5. What are the factors causing the negative test results?
A: the reasons in the Southern hybridization service can be summarized as follows:
(1) The abundance of target genes in the genome is lower than the detection limit of Southern blot hybridization experiment (the amount of target genes is lower than 0.1pg)
(2) In the state of incomplete genomic information of this species, it is difficult to ensure the specificity of the probe sequence, resulting in a large number of nonspecific bands.
(3) The quality of genome can not meet the requirements
(4) The sample to be tested is indeed negative
(5) Influence of enzyme digestion scheme
Seven. Service Process