One. Experimental Principle
SNP detection is a technology to detect the polymorphism of DNA sequence caused by the mutation of a single nucleotide in the chromosome genome. SNP exists in the form of single base transversion, transformation, insertion and deletion. As the third generation molecular marker, SNP marker has great development potential. It is widely used in many fields such as biology, agronomy, medicine and biological evolution. It plays an important role in molecular genetics, pharmacogenetics, forensic medicine, disease diagnosis and treatment.
Service Type:
1. Taqman Probe (Qualitative)
PCR primers and TaqMan probes is designed for different SNP sites on chromosomes for real-time fluorescent PCR amplification.
2. SNaPshot detection to SNP
It is mainly a typing technology based on the principle of fluorescent labeled single base extension, mainly for SNP typing projects with medium flux.
3. MassARRAy
Massarray mass spectrometry system is the most cost-effective medium and high-throughput SNP typing detection system in the market. DNA molecular fragments are excited by laser, and then the molecular weight of fragments is determined by flight time.
4. Illumina BeadXpress
Beadxpress system is used for batch SNP site detection, which can detect 1-384 SNP sites at the same time. And it is often used for genome chip result confirmation. It is suitable for high-throughput detection.
5. Direct sequencing
Among all SNP detection methods, direct amplification and sequencing of the fragments to be detected is the most accurate method, and it is also the gold standard of SNP analysis.
Two. Application Introduction
SNP can lead to human diseases, such as cancer, infectious diseases, autoimmune diseases, etc. SNP is an important basis for studying the genetic variation of human families, animals and plants. Therefore, SNP is widely used in the research of population genetics and disease-related genes, and plays an important role in pharmacogenomics, diagnostics and biomedical research.
Application fields:
1. Marking for genetic map;
2. Disease diagnosis and disease susceptibility genes identification;
3. Pharmacogenomics research and new drug screening ;
4. Drug metabolism and pharmacogenetics;
5. Forensic identification and individual identification;
6. Population genetics research and genetic polymorphism analysis;
7. Species identification, strain identification, etc.
Three. Experimental Methods
A. Direct sequencing:
a. Extract the genomic DNA of samples;
b. Designe and synthesize primers according to different regions;
c. Perform gene amplificationand pirification on all samples;
d. Sequencing;
e. Statistics and analysis.
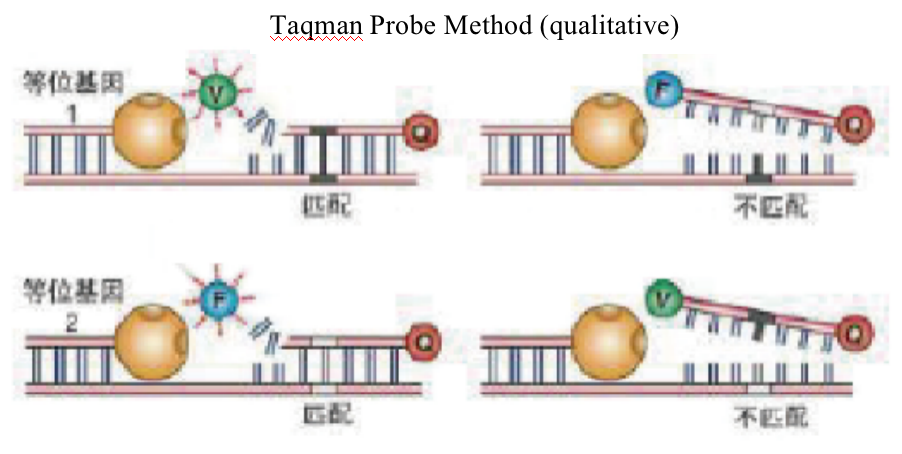
B. TaqMan probe method:
a. Extract genomic DNA of samples and carry out quality inspection;
b. Design and synthesize the probe;
c. Carry out pre experiment to optimze the PCR reaction system;
d. Realtime fluorescence quantitative PCR;
e. Analyze SNP locus data.
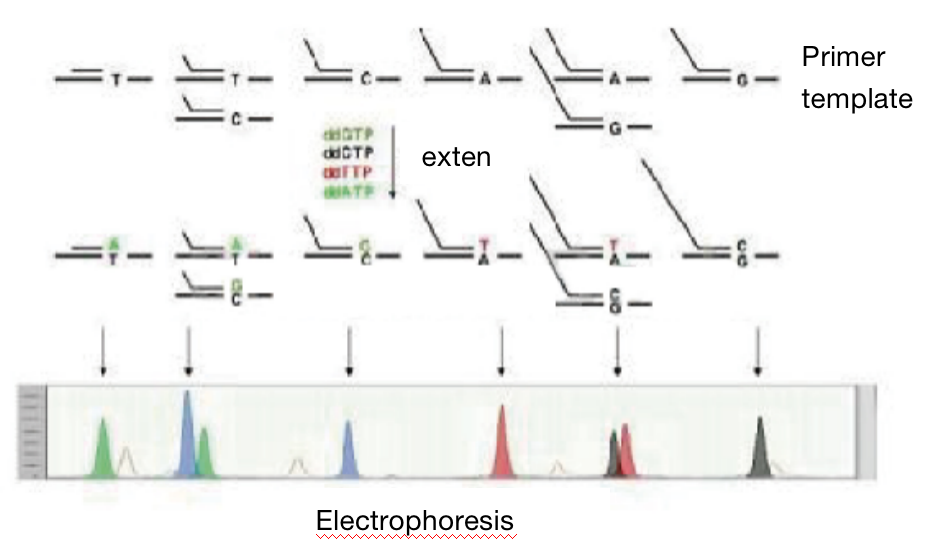
C. SNaPshot method:
a. Extract genomic DNA of samples and carry out quality inspection;
b. Design and synthesize the primers;
c. Amplification reaction;
d. Extension reaction;
e. Detect with sequencer;
f. Analyze the data.
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
Five. Case Display
Taqman Probe Method (qualitative)
SNaPshot method in testing SNP
![]()
Six.Common Problems
① In order to ensure the authenticity and reliability of the sequencing of the target area to be tested, the primer design should make the boundary of the target area to be tested at least 50bp away from the upstream and downstream primers;
② Online primer design is recommended to ensure the success rate;
③ In order to ensure the sequencing sensitivity, the fragment size of PCR product should be in the range of 250bp-650bp;
④ In order to facilitate the experiment, it is suggested that the primers should be assembled into one tube at the time of synthesis, so as to separate the PCR and sequencing primers;
⑤ In order to ensure the specificity of primers, it is recommended to confirm them on NCBI blast after primer design;
⑥ In order to prevent degradation, PCR products should be sequenced as soon as possible, otherwise they should be stored at - 20 ℃ and the time should not be too long;
⑦ In order to ensure the authenticity of the results, it is recommended to confirm the key points by reverse sequencing.
Seven. Service Process