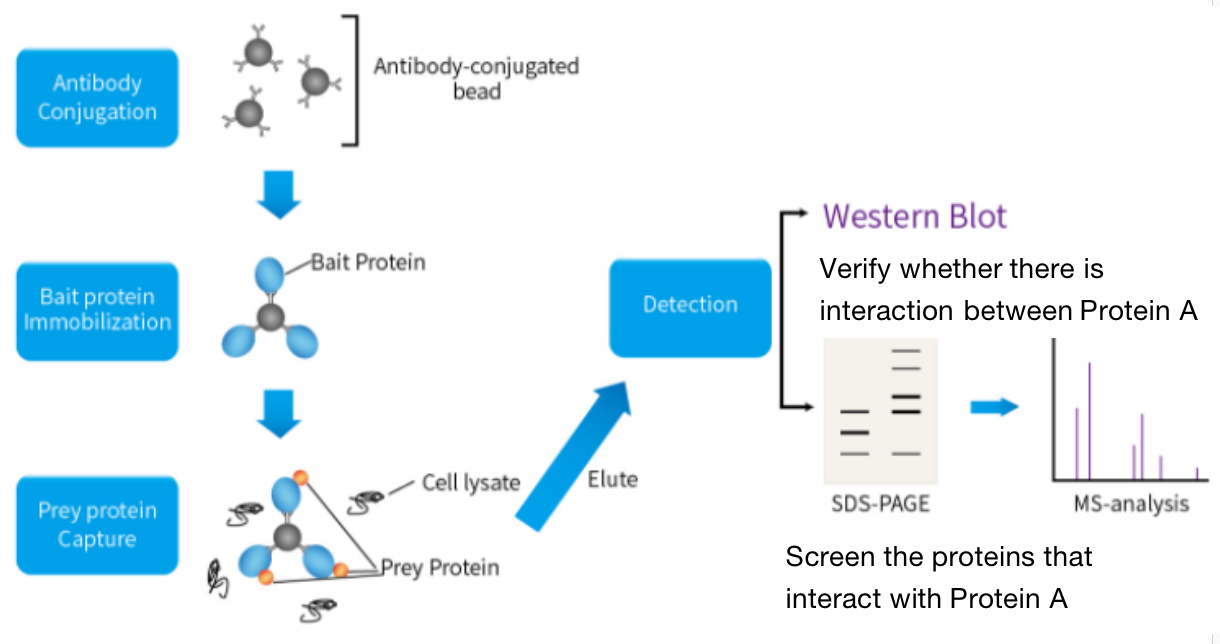
One. Experimental Principle
Immunocoprecipitation is a method that uses the specific binding of antigen and antibody and the specific binding of bacterial protein G or A to the Fc fragment of immunoglobulin. The basic principle is to add the antibody of the protein of interest to the cell lysate, and then add protein G or A specifically bound to the antibody and bound to the magnetic beads after incubation. If there is a target protein bound to the protein of interest in the cell, a protein complex can be formed: "target protein - protein of interest - anti-interest protein antibody - protein G or A"; Finally, the target protein is detected by immunoblotting.
When there is cell lysis under non denaturing conditions, many protein-protein interactions existing in intact cells are retained. If X is immunoprecipitated with an antibody to protein X, protein Y bound to X in the body can also be precipitated. Co-IP determines that there is an interaction relationship, but this interaction does not determine whether to use direct or indirect interaction.
1) Advantages of immunoprecipitation
1. The interacting proteins are all post-translational modified and in a natural state.
2. The interaction between proteins is carried out in a natural state, which can avoid human influence.
3. Interacting protein complexes in natural state can be isolated.
2) Disadvantages of immunoprecipitation
1. Low affinity and transient protein-protein interaction may not be detected;
2. The binding of the two proteins may not be direct, but a third party may act as a bridge between them;
3. It is necessary to predict what the target protein is before the experiment to select the antibody to be tested finally. Therefore, if the prediction is incorrect, the experiment will not get the result. The method itself is risky.
Two. Application Introduction
As common molecules, Co-IP, ChIP, RIP, and Pulldown act as verification methods for each other. They can be distinguished from the starting point of the test: co-IP, ChIP, and RIP use known proteins to detect related proteins, DNA and RNA in the body, and separate the target proteins, DNA and RNA by protein antibody immunoprecipitation. Pulldown is to test related proteins in vitro according to known RNA (RNA pulldown) or known protein (GST pulldown).
Co-IP is mainly used to detect the interaction between proteins. It is an extension of IP experiment. Based on the potential of IP reaction, it can capture and purify the main targets (antigens, etc.) and target other bound macromolecules through natural interactio in the sample solution. In addition, Co-IP can also be used to determine protein interactions under different conditions.
Three. Experimental Method
Four. Sample Delivering Requirements
Sample type | Sample requirements | Preservation conditions | Delivery conditions | Note |
Animal tissue | A single sample should be less than 0.1g, and installed in 2ml EP tube or frozen storage tube, and do not overdose it; Samples should be as fresh as possible. If protein or nucleic acid cannot be extracted immediately, they should be frozen at - 80 ℃ or lower after quick freezing with liquid nitrogen. Frozen samples should avoid repeated freezing and thawing to avoid degradation. | At - 80 ℃ | With dry ice | All samples need to be uniquely marked and the markings are clearly identifiable |
Seed sample | A single sample of seed that are shelled and fresh or stored in liquid nitrogen should be less than 0.2g | At - 80 ℃ | With dry ice | |
Adherent/Suspension cell | 1. Total RNA or protein: 10^5 cell/index, plasma/ nuclear RNA or protein:10^7 cell/index, mitochondrial RNA or protein: 2*10^7 cell/index.the samples should be as fresh as possible and should directly add Trizol (QPCR test) or frozen at - 80 ℃ after collected. 2. If the cells are in poor condition after treatment (dosing, transfection and infection), the sample collection volume should be increased as appropriate | At - 80 ℃ | With dry ice | |
Whole blood/serum samples | 5 to 10 ml of peripheral blood and 1 to 3 ml of bone marrow preserved with anticoagulant tubes. The leukocyte homogenate stored at -80 ℃ for no more than half a week is less than 400 μ L, and 400 μ L Trizol is added every 400 μ L | At - 80 ℃ | With dry ice | |
Paraffin embedded samples | The effective thickness of paraffin block embedded by standard paraffin embedding box should be thicker than 0.1cm; The thickness of each fresh FFPE tissue sections should be no more than 10 microns, and a surface area of no more than 250 mm^2, 2 to 8 piece in total. | At - 20℃ | With ice bag | |
Antibody | 1. Provide antibodies that meet the corresponding experimental requirements according to the sample species; 2. Send according to the requirements of the antibody manual, and try to avoid sub packaging; In case of sub packaging, ensure that the amount is sufficient for the experiment, and provide the antibody instruction; 3. The antibody tube storing the antibody should have a mark that can recognize the antibody, and the amount of antibody should be greater than the amount required for the experiment. | At - 20℃ | With ice bag | |
Primers | The primers should be dry powder and less than or equal to 1 OD. If pre experiment is conducted, at least two pairs of primers should be provided for each gene, and the primer synthesis sheet should be attached by the company, | At - 20℃ | With ice bag or at ambient temperature |
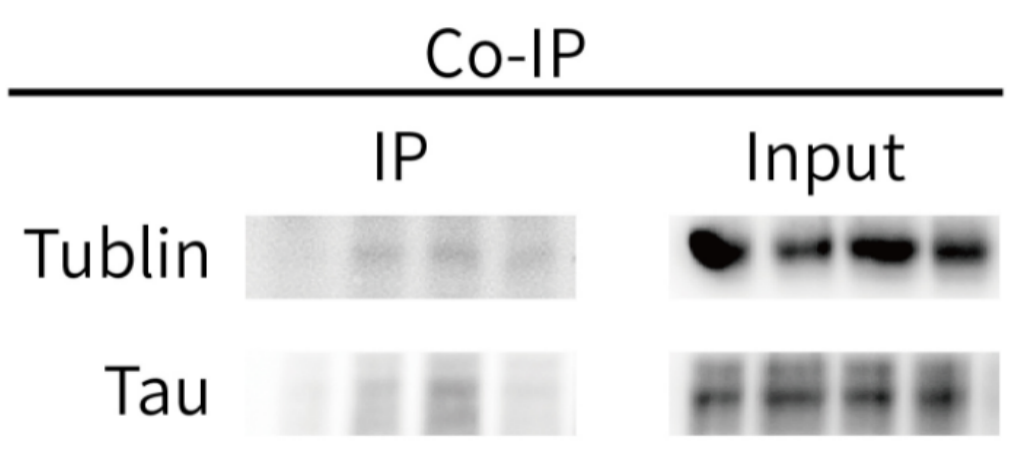
Five. Case Display
Six. Common Problems
1. Preparation of experimental sample preparation method: according to the research purpose, the test sample may be tissue sample or cell sample. The preparation processes of different samples are different, and the expression of target protein is also quite different. Appropriate samples and sample preparation methods must be selected.
2. Select the appropriate specific antibody of protein according to the research purpose: the antibody used for Co-IP experiment is the specific antibody against the target protein. The most ideal is to select an antibody that can recognize an epitope of the protein of interest, and this epitope will not be masked by the interaction of any other protein, which the requirement is much higher than that of ordinary WB antibody; At the same time, only when the specificity and sensitivity of antibody are as high as possible can there be a clean background and better IP effect. Therefore, the most suitable antibody must be selected for CO IP experiment.
3. No target protein or few proteins are detected:
Possible Cause | Solution |
The sample is degraded by protease | Add protease inhibitor; Perform all operations on ice and keep the temperature within 4°C and prevent from freezing and thawing. |
The concentration of the antibody is too low. | Adjust the concentration of IP and/or IB antibody, set up concentration gradient to explore the best concentration if necessary. |
The affinity of anti-antibody is too low | Select the corresponding antibody suitable for IP and/or IB |
The IP antibody is not bind to agarose beads. | Select the corresponding beads suitable for IP and store them correctly to prevent deterioration or drying |
Tag is not exposed to the surface of the fusion protein conformation | Change the expression site of tag |
The stringency of the lysis solution is too high | Replace it with the lysis solution with low-stringency |
4.The target protein presents high background
Possible Cause | Solution |
There is nonspecific protein binding. | Lyse cells in serum-free medium; Pre-wash with protein (G/A) beads before immunoprecipitation, and increase rinsing times and preciseness (high salt or detergent) after immunoprecipitation |
The stringency of the lysis solution is too low | Replace it with the lysis solution with high-stringency |
The experimental instrument or liquid is contaminated | Use the cleaned experimental instrument or liquid |
There is nonspecific adsorption on transfer membrane | Wear gloves and clamp with tweezers. Do not touch the membrane transfer surface |
Seven. Sevice Process